A new high-throughput method to assess risk from pesticide co-exposures
Honey bees are almost always exposed to pesticides. For example, in a recent survey of 198 beekeeper hives throughout New York, we found pesticides in comb wax from 197 of the hives (Wheeler et al. 2018). On average, wax from the hives contained 6 different pesticides. And this is actually low pesticide exposure compared to some contexts. For example, when we screened bee bread from colonies conducting New York apple pollination in 2019, we found an average of 14 pesticides, with bee bread from some hives containing 23 different pesticides (Zhao et al., in prep).
The fact that pesticide co-exposures are so common poses a major problem for risk assessment by regulatory agencies such as the U.S. Environmental Protection Agency. Those agencies must determine whether risk from particular pesticides is high enough to warrant restrictions on their use. But currently, the EPA and other regulatory agencies only consider risk from individual pesticides, not combinations. This gives an incomplete picture of risk since 1) co-exposures to bees are ubiquitous, and 2) we know that some pesticides interact with each other (i.e., synergize) such that the blend is much more toxic than the individual pesticides on their own.
But how can we possibly get a handle on risk from co-exposures when honey bees potentially encounter hundreds of different pesticides? Some quick math shows that if there are 100 different pesticides that a honey bee might encounter, we’d need to screen 4,950 pesticide combinations to understand risk from dual exposures. If we want to assess risk from combinations of 3-pesticide exposures, that’s 161,700 assays. And what about risk from combinations of 14 different pesticides, which is the average number of pesticides we found in bee bread from hives doing New York apple pollination? That’s 44,186,942,677,323,600 assays. Good luck!
Clearly we need a quick and efficient way to screen honey bees for risk from pesticide combinations if we’re going to start to get a handle on this important issue. This is the topic for our thirty-ninth Notes from the Lab, where we summarize “Pesticide risk assessment at the molecular level using honey bee cytochrome P450 enzymes: A complementary approach,” written by Julian Haas and Ralf Nauen and published in Environment International [2021, 147:106372].
For their study, Haas and Nauen (Photo 1) took an approach that’s similar to how drugs for humans are initially screened in the lab to determine whether drug-drug interactions have the potential to occur. Specifically, they assessed how well honey bee detoxification enzymes performed at breaking down pesticides into non-toxic products using 384-well microplates (Photo 2). Once they knew their assays were working well, they then tested the same pesticides on live honey bees and compared their high-throughput microplate assay results to what happened with the bees.
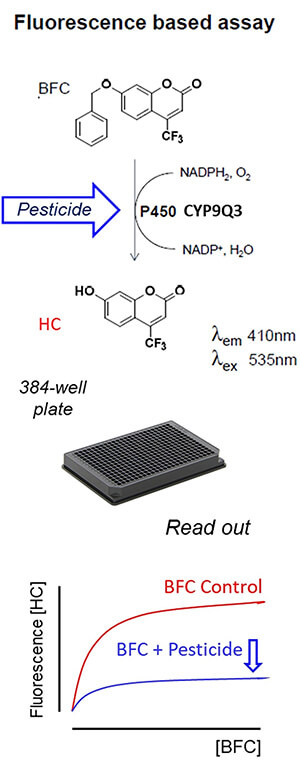
The basic concept of the microplate assay is shown in Figure 1. There are a lot of acronyms and symbols in the figure, but bear with me — it’s actually pretty easy to explain and provides a nice overview of how the authors’ microplate assay works.
Starting at the top, the important point is that BFC (the molecule at the top) can potentially be broken down to HC (the molecule below the arrow) in the presence of the detoxification enzyme P450 CYP9Q3. However, this enzyme isn’t able to convert as much BFC to HC in the presence of some pesticides (shown via the blue arrow). This could be for either of two reasons. First, the pesticide may compete with BFC for access to the detoxification enzyme (because it is also detoxified by that enzyme!). Such is the case for the neonicotinoid insecticides thiacloprid and acetamiprid, which are detoxified by CYP9Q3 in honey bees. Alternatively, a pesticide may inhibit the functioning of the enzyme, making it less efficient at detoxifying other things. As we will see later, such is the case for some fungicides, which inhibit CYP9Q3 and therefore interfere with the ability of honey bees to detoxify thiacloprid and acetamiprid.
Now move to the figure below the picture of the 384-well plate. This is what the authors monitored in the microplate assays: the amount of HC produced (measured via fluorescence) as more BFC was added in the presence of the CYP9Q3 detoxification enzyme. The blue curve is below the red curve, meaning less HC was produced in the presence of the pesticide. This indicates the pesticide either competes with or inhibits the CYP9Q3 detoxification enzyme.
OK, now that we’re all on the same page with the authors’ slick microplate assay, let’s get to what they tested. First, Haas and Nauen assessed whether honey bee detoxification enzyme CYP9Q3 showed evidence of detoxifying multiple neonicotinoid insecticides (thiacloprid, acetamiprid, imidacloprid, thiamethoxam) and a pyrethroid insecticide (tau-fluvalinate, which is also used as varroacide). Next, they tested whether several different fungicides (triflumizole, propiconazole, triadimefon, epoxiconazole, uniconazole, prothioconazole, prochloraz, and azoxystrobin) interfered with the detoxification enzyme. Finally, the authors performed LD50 studies with two of the neonicotinoids (thiacloprid and acetamiprid) on their own or via co-exposure with the fungicides. These LD50 bioassay data were then directly compared to the microplate enzyme activity results.
So, what did they find? Did the microplate assay show that insecticides are broken down by the CYP9Q3 detoxification enzyme? Yes and no. Haas and Nauen found evidence that ….


